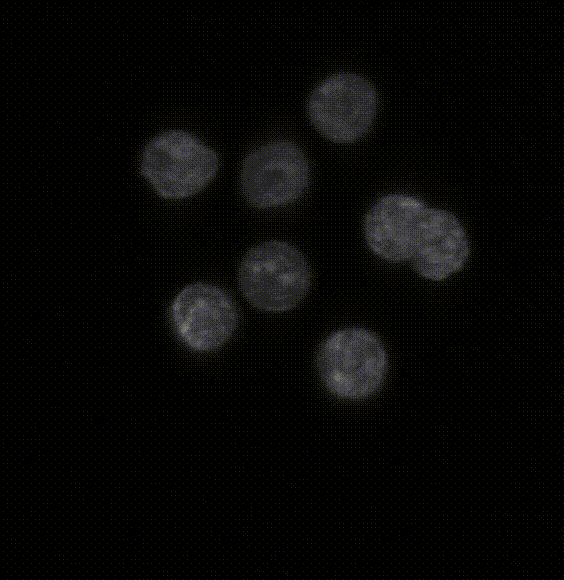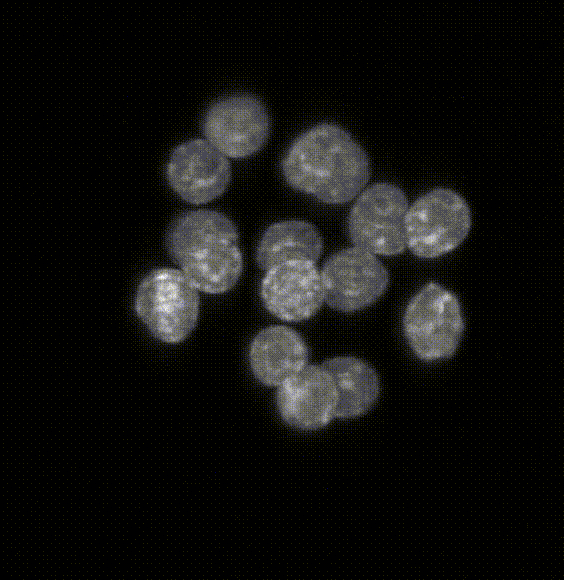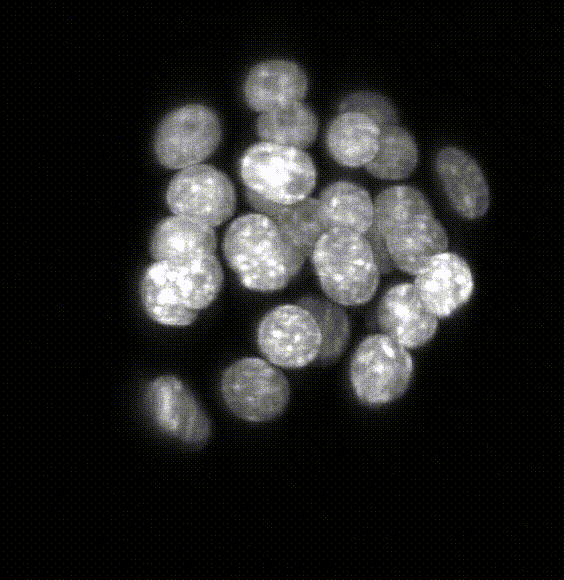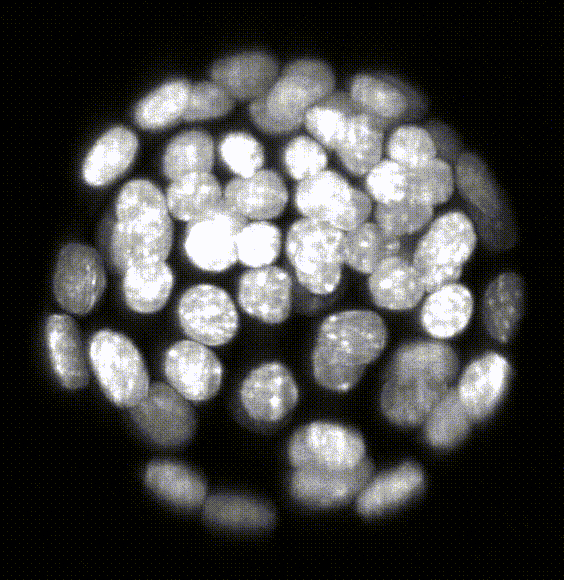
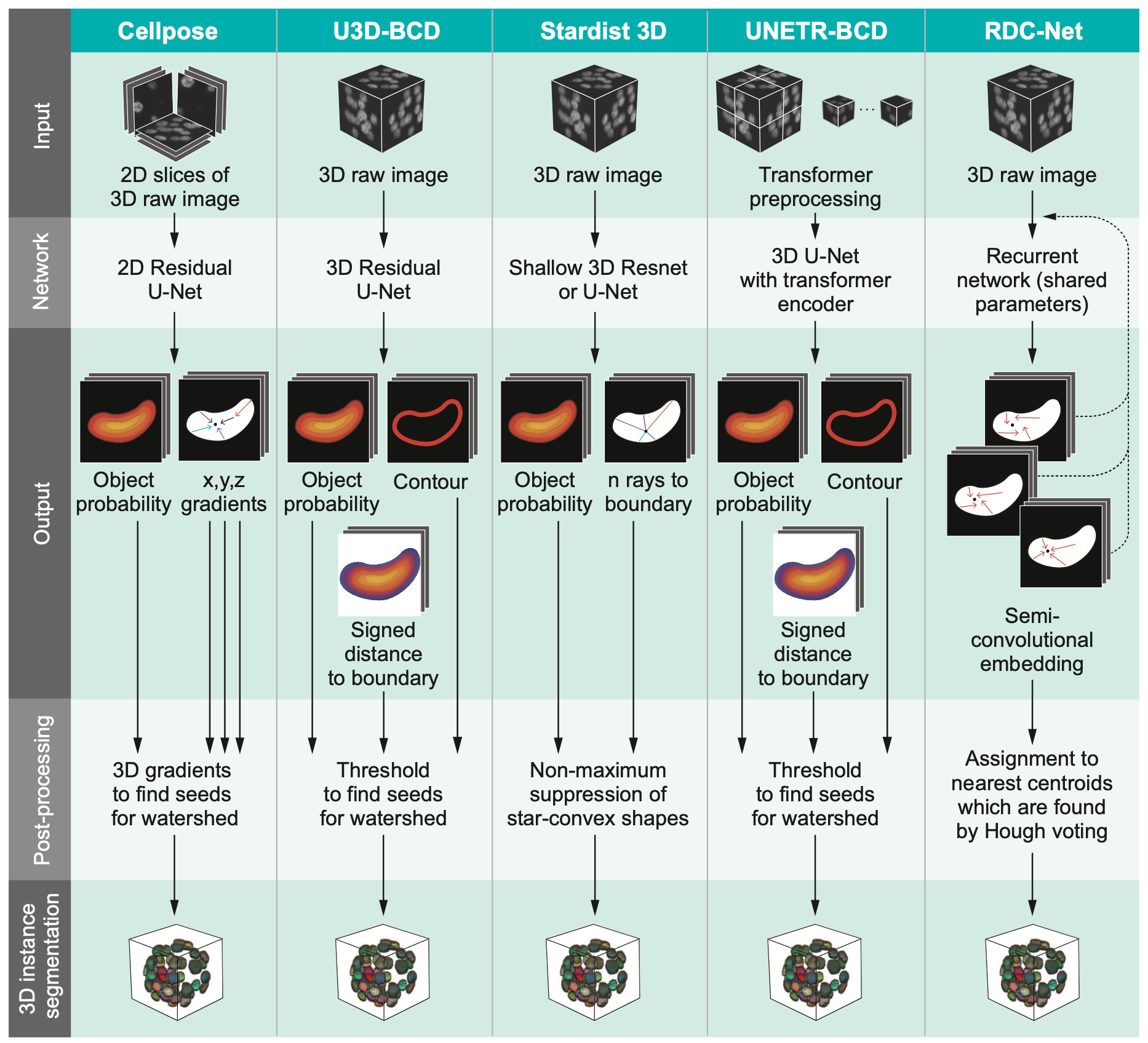
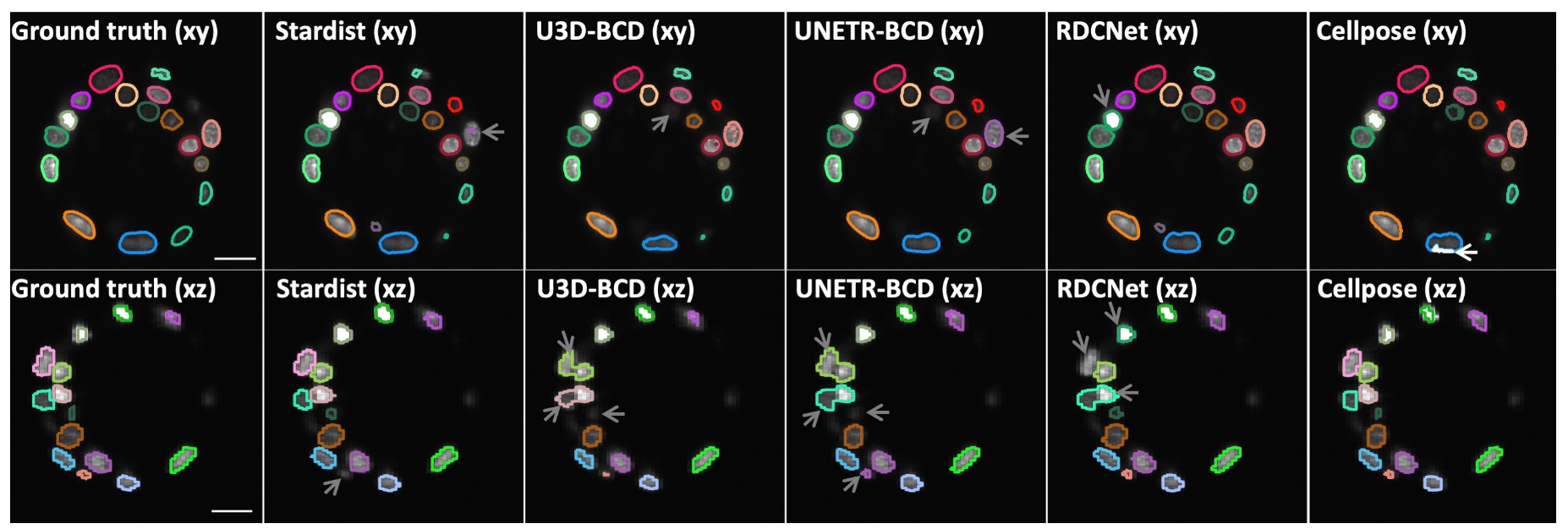
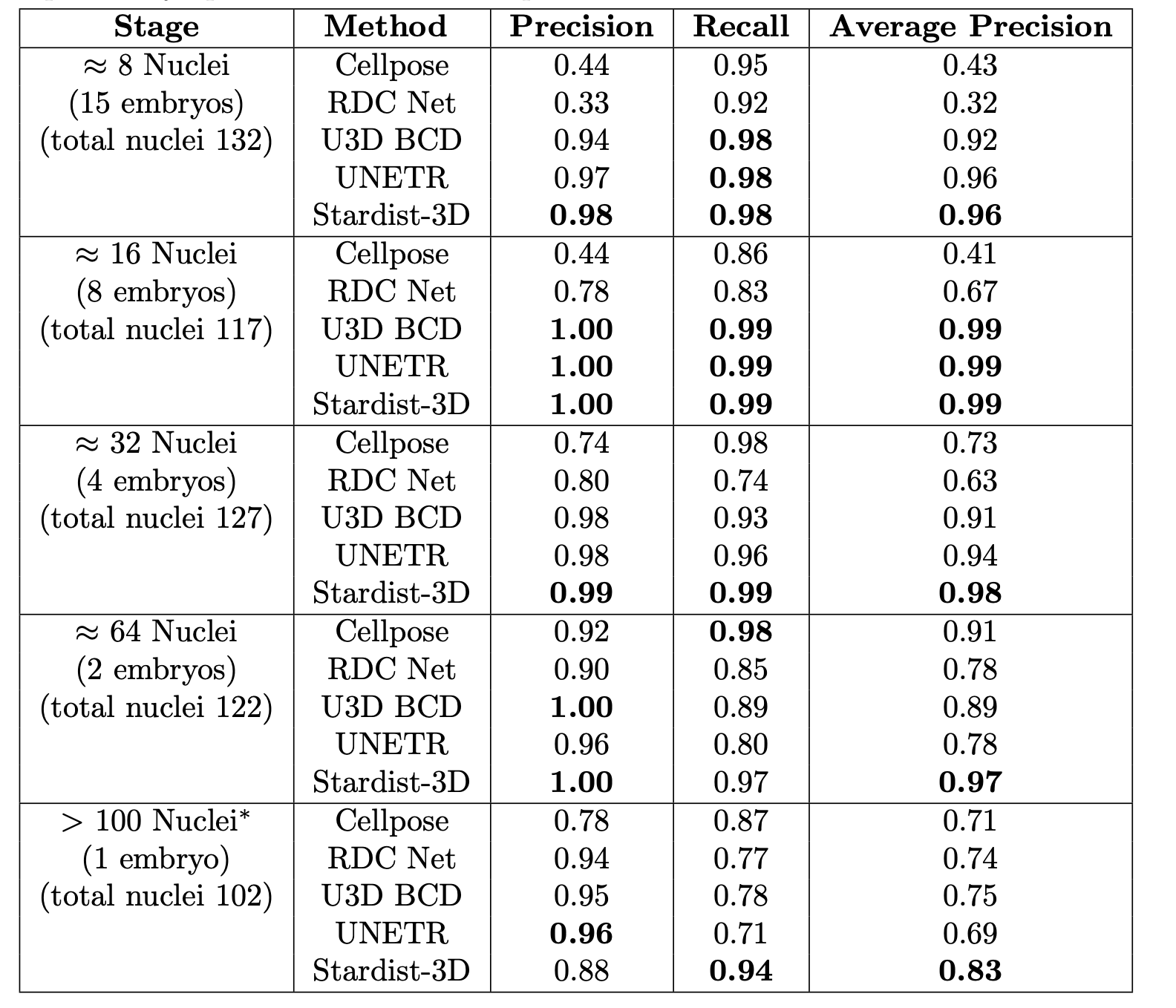
Comparison of five deep-learning-based methods for nuclear instance segmentation
In the illustration to the left each column depicts a different method. From top to bottom, the rows illustrate how images are inputted into the network, the network’s architecture, network outputs for a single nucleus, the post-processing steps, and the 3D instance segmentation, respectively.
Click the image for a detailed view.